Implement a Topic-specific Q&A Application with LangChain and Oracle GenAI

Now that I caught your attention, I hope the image isn't too triggering. I grew up learning to fear these little black monsters that potentially carry a load of Dengue virus that could make you really sick. As recently reported, the disease continues to be an issue in Singapore. During my undergraduate years, I was deeply passionate about viral diseases, and spent a few months in the laboratory learning to characterize the structural components of the virus using cyrogenic electron microscopy. Hence, when Luc Demanche challenged me again to demonstrate retrieval-augmented generation (RAG), I decided to collect and use a few recent articles (PMIDs 38140548, 37478848, 37114191), and use them to create a quick proof-of-concept Oracle APEX application that allows the user to ask questions about the disease. These articles cover topics on the Dengue virus, its genomics and ultrastructure, and progress in vaccine development.
The Ingredients
In my last post, I had described how LangChain could be used to quickly create AI applications using the power of a large language modal (LLM). The next ingredient needed was an approach to deploy these models and expose an API so that these models can be used for performing inferences. Since my goal was to use Oracle APEX as the frontend application, turning the LangChain application into a web service was required.
As mentioned, newer versions of Oracle Accelerated Data Science would allow us to deploy LangChain applications to the Oracle Cloud Infrastructure (OCI) Data Science Service (ODS), that along with Jupyter notebooks and pipelines, allows data scientists to manage and deploy ML models on managed infrastructure. Unfortunately, I had hit a roadblock and am waiting to resolve the issue with Oracle Support.
Meanwhile, I looked at the next best thing, and that is, LangServe. LangServe makes it very convenient to write LangChain applications and make them available as an API. It builds on top of FastAPI, a Python-based web framework. When deployed, the endpoints for invoking the applications, API docs, and a playground are created automatically. However, like LangChain, its still a product in its infancy.
The PDF documents, for good reason, are uploaded to an Object Storage bucket. This bucket is then mounted on the Compute where LangServe is installed. To mount the bucket as a file system, I used the s3fs-fuse utility. You can read more about that integration here. There are other LangChain document loaders to be explored, which I'll do at a later time.
An Oracle Autonomous Database hosts the Oracle APEX application. Not wanting to fuss around with how to make this endpoint available to an Oracle Autonomous Database securely, I went with running LangServe on a compute situated in a private subnet, and then expose the endpoints using the OCI API Gateway.
Here's a simple topology to help visualise the interaction between the various OCI components described above.
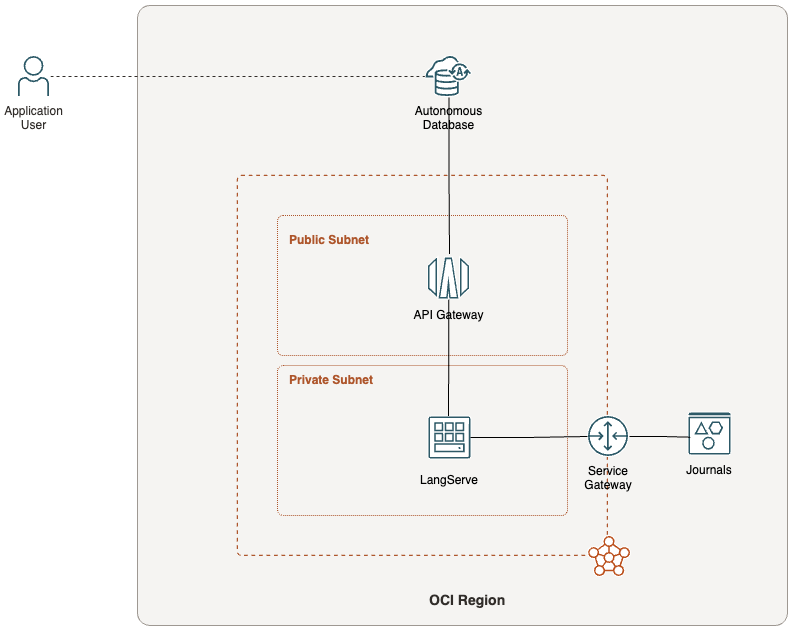
The Recipe
To create the environment for the Compute, I used Conda to create the environment using the following configuration:
channels:
- defaults
- conda-forge
dependencies:
- python=3.11
- langchain=0.1.13
- langchain-community=0.0.29
- faiss=1.7.4
- pypdf=3.17.4
- langchain-text-splitters=0.0.1
- pip
- pip:
- oci==2.125.0
- oci-cli==3.38.0
- databricks-cli==0.18.0
- mlflow==2.11.3
- oracle-ads[data,notebook,torch]==2.11.5
- langserve[server]==0.0.51
- langchain-cli==0.0.21
Note
I had created the environment file for an ODS notebook. You could omit the PIP installs for the OCI and Oracle ADS packages.
I'm still very much a noob, so the code follows the instructions found in the LangChain tutorial on RAG. We begin by declaring the import statements.
from fastapi import FastAPI
from langchain_community.vectorstores import FAISS
from langchain_community.embeddings import OCIGenAIEmbeddings
from langchain_community.llms import OCIGenAI
from langchain_community.document_loaders import PyPDFDirectoryLoader
from langchain_core.prompts import PromptTemplate
from langchain_core.output_parsers import StrOutputParser
from langchain.schema.runnable import RunnablePassthrough
from langchain_text_splitters import RecursiveCharacterTextSplitter
from langserve import add_routes
The first task in this Python application is to load the PDFs into a list of Document objects. However, since these documents tend to be large, they will need to be sliced into smaller chunks. To do that, I used a text splitter, and load all the PDF files at once.
text_splitter = RecursiveCharacterTextSplitter(
chunk_size=3000,
chunk_overlap=50,
length_function=len,
is_separator_regex=False,
)
loader = PyPDFDirectoryLoader("/mnt/journals")
docs = loader.load_and_split(text_splitter=text_splitter)
Next, instantiate the embeddings model. We will use the OCI GenAI service that currently only supports Cohere's embedding models.
embeddings = OCIGenAIEmbeddings(
auth_type="API_KEY", # Or, preferably, use RESOURCE_PRINCIPAL
auth_profile="DEFAULT", # Omit if using RESOURCE_PRINCIPAL
model_id="cohere.embed-english-v3.0",
service_endpoint="https://inference.generativeai.us-chicago-1.oci.oraclecloud.com",
compartment_id="ocid1.compartment.oc1..."
)
For the Vector store, we will use Meta's FAISS as described in the cookbook, and unglamourously store the vectors in memory. Did I say that this is a PoC? 😉 Use the wrapper function from_documents to create the vector store using the loaded list of documents, and then create the retriever object.
vectorstore = FAISS.from_documents(
documents=docs,
embedding=embeddings
)
retriever = vectorstore.as_retriever()
Note
This is not a production-ready solution. Keep in mind that, as the documentation explains, the wrapper function is intended for, I quote, "a quick way to get started". It embeds the documents provided, creates the in-memory docstore, and then initialises the FAISS database.
There is a severe limitation to this wrapper function. When calling the OCI GenAI EmbedText endpoint, the inputs array is limited to a maximum of 96 strings, and each string is limited to 512 tokens. By default, the
truncateargument is set toEND. When chunking the documents, I had to find a balance to make sure the chunks are small enough not to generate more than 96 document objects. This allows me to create the vector store with a very limited number of PDF files. So, in a real world implementation, I wouldn't use the wrapper function, and of course, a better docstore.
Prepare the prompt.
template = """Answer the question based on the documents provided:
{context}
Question: {question}
"""
prompt = PromptTemplate.from_template(template)
Note
This prompt seemed to help avoid having the assistant respond to questions that are outside the given context.
Then instantiate the LLM that will be used for the text generation. Here, I used the OCI GenAI's Cohere command model.
llm = OCIGenAI(
auth_type="API_KEY", # Or, preferably, use RESOURCE_PRINCIPAL
auth_profile="DEFAULT", # Omit if using RESOURCE_PRINCIPAL
model_id="cohere.command",
service_endpoint="https://inference.generativeai.us-chicago-1.oci.oraclecloud.com",
compartment_id="ocid1.compartment.oc1...",
model_kwargs={"temperature": 0, "max_tokens": 500}
)
Assemble the chain.
chain = (
{"context": retriever, "question": RunnablePassthrough()}
| prompt
| llm
| StrOutputParser()
)
Create the FastAPI application, and then add a route using the chain.
app = FastAPI(
title="Ask the Expert",
version="1.0",
description="A demo RAG system for answering questions based on a collection of biomedical publications.",
)
add_routes(
app,
chain,
path="/asktheexpert"
)
Finally, run the application in a Uvicorn web server:
if __name__ == "__main__":
import uvicorn
uvicorn.run(app, host="0.0.0.0", port=8000)
The Oracle Autonomous Database has strict rules on what remote endpoints it is willing to communicate with.
- The endpoint must be resolvable, so no IP addresses.
- The endpoint must be secured using SSL/TLS, and the certificates must be verifiable.
Using an API Gateway to front the LangServe instance would meet these requirements. The endpoint URL to invoke the LangChain application would something like this:
https://{{UNIQUE_STRING}}.apigateway.{{REGION}}.oci.customer-oci.com/demo/v1/asktheexpert/invoke
This endpoint takes the question embedded in a JSON with the following schema:
{
"input": "What is the role of the NS3 protein?"
}
A successful invocation of the API will respond with results in JSON.
{
"output": " The NS3 protein is a serine protease and ...",
"callback_events": [],
"metadata": {
"run_id": "f2f3dac3-1391-4f34-84ce-858a591adc74"
}
}
Invoking the LangChain application to get your questions answered within an Oracle APEX application is to then use the PL/SQL API, APEX_WEB_SERVICE, as you would with any other web service.
declare
l_response clob;
l_response_json json_object_t;
begin
apex_web_service.set_request_headers(
p_name_01 => 'Content-Type'
, p_value_01 => 'application/json'
);
l_response := apex_web_service.make_rest_request(
p_http_method => 'POST'
, p_url => 'https://{{UNIQUE_STRING}}.apigateway.{{REGION}}.oci.customer-oci.com/demo/v1/asktheexpert/invoke'
, p_body => json_object('input' value apex_escape.json(:P1_QUESTION))
);
if apex_web_service.g_status_code = 200 then
l_response_json := json_object_t.parse(l_response);
:P1_RESPONSE := l_response_json.get_string('output');
else
raise_application_error(-20000, 'Failed to get a response.');
end if;
end;
Here's the quick demo in action.
Closing Words
This is one of many approaches for implementing a RAG solution to query structured and unstructured data that matters to you. Again, this blog post is simply to outline what's possible that I hope will provide you a head start. If you'd like to discuss your use cases for AI or machine learning, please do not hesitate to reach out.
Credits
Photo by National Institute of Allergy and Infectious Diseases on Unsplash
